

Next: About this document ...
Up: unfolding_distributions
Previous: 4 Double-integral Kramers' theory
-
G. I. Bell.
- Models for the specific adhesion of cells to cells.
Science, 200 (4342): 618-627, May 1978.
ISSN 0036-8075.
URL http://www.jstor.org/stable/1746930.
The Bell model and a fair bit of cell bonding background.
-
Elena Braverman and Reneeta Mamdani.
- Continuous versus pulse harvesting for population models in constant
and variable environment.
J Math Biol, 57 (3): 413-434, September
2008.
ISSN 0303-6812.
doi: rm10.1007/s00285-008-0169-z.
URL http://www.springerlink.com/content/a1m23v50201m2401/.
An example of non-exponential Gomperz law.
-
Mariano Carrion-Vazquez, Andres F. Oberhauser, Susan B. Fowler, Piotr E.
Marszalek, Sheldon E. Broedel, Jane Clarke, and Julio M. Fernandez.
- Mechanical and chemical unfolding of a single protein: A comparison.
Proceedings of the National Academy of Sciences, 96
(7): 3694-3699, 1999.
doi: rm10.1073/pnas.96.7.3694.
URL http://www.pnas.org/cgi/content/abstract/96/7/3694.
-
O. K. Dudko, A. E. Filippov, J. Klafter, and M. Urbakh.
- Beyond the conventional description of dynamic force spectroscopy of
adhesion bonds.
Proc Natl Acad Sci U S A, 100 (20):
11378-11381, September 2003.
ISSN 0027-8424.
doi: rm10.1073/pnas.1534554100.
URL http://www.pnas.org/content/100/20/11378.abstract.
-
Olga K. Dudko, Gerhard Hummer, and Attila Szabo.
- Intrinsic rates and activation free energies from single-molecule
pulling experiments.
Phys Rev Lett, 96 (10): 108101, March 2006.
ISSN 0031-9007.
doi: rm10.1103/PhysRevLett.96.108101.
-
Olga K. Dudko, Jérôme Mathé, Attila Szabo, Amit Meller, and Gerhard
Hummer.
- Extracting kinetics from single-molecule force spectroscopy: nanopore
unzipping of DNA hairpins.
Biophys J, 92 (12): 4188-4195, June 2007.
ISSN 0006-3495.
doi: rm10.1529/biophysj.106.102855.
-
E. Evans and K. Ritchie.
- Dynamic strength of molecular adhesion bonds.
Biophys J, 72 (4): 1541-1555, April 1997.
ISSN 0006-3495.
URL http://www.biophysj.org/cgi/content/abstract/72/4/1541.
-
E. Evans and K. Ritchie.
- Strength of a weak bond connecting flexible polymer chains.
Biophys J, 76 (5): 2439-2447, May 1999.
ISSN 0006-3495.
URL http://www.biophysj.org/cgi/content/abstract/76/5/2439.
Develops Kramers improvement on Bell model for domain unfolding.
Presents unfolding under variable loading rates. Often cited as the
``Bell-Evans'' model? They derive a unitless treatment, scaling force by
 , TODO; time by
, TODO; time by 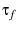 , TODO; elasiticity by compliance
, TODO; elasiticity by compliance 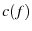 .
The appendix has relaxation time formulas for WLC and FJC polymer models.
.
The appendix has relaxation time formulas for WLC and FJC polymer models.
-
Benjamin Gompertz.
- On the nature of the function expressive of the law of human
mortality, and on a new mode of determining the value of life contingencies.
Philosophical Transactions of the Royal Society of London,
115: 513-583, 1825.
ISSN 02610523.
URL http://www.jstor.org/stable/107756.
-
Peter Hänggi, Peter Talkner, and Michal Borkovec.
- Reaction-rate theory: fifty years after kramers.
Rev. Mod. Phys., 62 (2): 251-341, Apr 1990.
doi: rm10.1103/RevModPhys.62.251.
URL http://prola.aps.org/abstract/RMP/v62/i2/p251_1.
The Kramers' theory review article. See pages 268-279 for the
Kramers-specific introduction.
-
Gerhard Hummer and Attila Szabo.
- Kinetics from nonequilibrium single-molecule pulling experiments.
Biophys J, 85 (1): 5-15, July 2003.
ISSN 0006-3495.
URL http://www.biophysj.org/cgi/content/abstract/85/1/5.
READ.
-
Changbong Hyeon and D. Thirumalai.
- Can energy landscape roughness of proteins and RNA be measured by
using mechanical unfolding experiments?
Proc Natl Acad Sci U S A, 100 (18):
10249-10253, September 2003.
ISSN 0027-8424.
doi: rm10.1073/pnas.1833310100.
URL http://www.pnas.org/cgi/content/abstract/100/18/10249.
Derives the major theory behind my thesis. The Kramers rate equation
is Hanggi Eq. 4.56c (page 275)Hänggi et al. (1990).
-
S. Izrailev, S. Stepaniants, M. Balsera, Y. Oono, and K. Schulten.
- Molecular dynamics study of unbinding of the avidin-biotin complex.
Biophys J, 72 (4): 1568-1581, April 1997.
ISSN 0006-3495.
URL http://www.biophysj.org/cgi/content/abstract/72/4/1568.
-
D. A. Juckett and B. Rosenberg.
- Comparison of the gompertz and weibull functions as descriptors for
human mortality distributions and their intersections.
Mech Ageing Dev, 69 (1-2): 1-31, June 1993.
ISSN 0047-6374.
doi: rm10.1016/0047-6374(93)90068-3.
-
S. J. Olshansky and B. A. Carnes.
- Ever since gompertz.
Demography, 34 (1): 1-15, February 1997.
ISSN 0070-3370.
URL http://www.jstor.org/stable/2061656.
-
Michael Schlierf and Matthias Rief.
- Single-molecule unfolding force distributions reveal a funnel-shaped
energy landscape.
Biophys J, 90 (4): L33-L35, February 2006.
ISSN 0006-3495.
doi: rm10.1529/biophysj.105.077982.
URL http://www.biophysj.org/cgi/content/abstract/90/4/L33.
The inspiration behind my sawtooth simulation. Bell model fit to
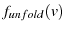 , but Kramers model fit to unfolding distribution for a given
, but Kramers model fit to unfolding distribution for a given
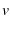 . Eqn. 3 in the supplement is Evans-Ritchie 1999's Eqn. 2Evans and Ritchie (1999),
but it is just ``[dying percent] * [surviving population] = [deaths]'' (TODO,
check).
. Eqn. 3 in the supplement is Evans-Ritchie 1999's Eqn. 2Evans and Ritchie (1999),
but it is just ``[dying percent] * [surviving population] = [deaths]'' (TODO,
check).
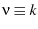 is the force/time-dependent off rate... (TODO) The
Kramers' rate equation (second equation in the paper) is Hanggi Eq. 4.56b
(page 275)Hänggi et al. (1990). It is important to extract
is the force/time-dependent off rate... (TODO) The
Kramers' rate equation (second equation in the paper) is Hanggi Eq. 4.56b
(page 275)Hänggi et al. (1990). It is important to extract 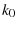 and
and 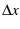 using every available method.
using every available method.
-
N.G. van Kampen.
- Stochastic Processes in Physics and Chemistry.
Elsevier, North-Holland Personal Library, Amsterdam, 2007.
-
Jong-Wuu Wu, Wen-Liang Hung, and Chih-Hui Tsai.
- Estimation of parameters of the Gompertz distribution using the
least squares method.
Applied Mathematics and Computation, 158
(1): 133-147, October 2004.
ISSN 0096-3003.
doi: rm10.1016/j.amc.2003.08.086.
URL
http://www.sciencedirect.com/science/article/B6TY8-4B3NR1W-B/1/bbaa47878ada03c6ef8e681d03bb65d3.


Next: About this document ...
Up: unfolding_distributions
Previous: 4 Double-integral Kramers' theory
Download unfolding_distributions.pdf
View the source files or my
.latex2html-init configuration file
<#1601#>Bibliography<#1601#>
Copyright © 2009-10-12, W. Trevor King
(contact)
Released under the GNU Free Document License, Version 1.2 or later
Drexel Physics