Open source
single molecule
force spectroscopy
Protein unfolding in varying salt concentrations
Trevor King
Open source SMFS
Proteins: What are they?
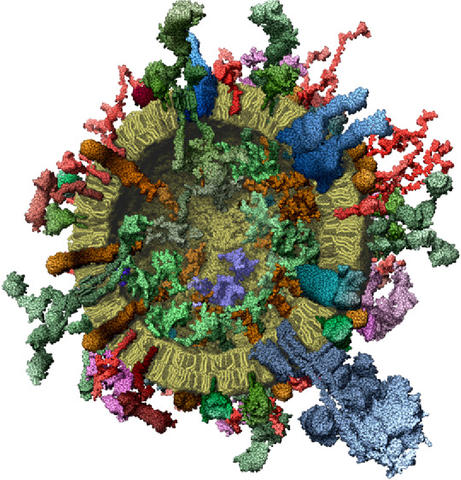
Proteins: Where are they?
Proteins: Titin
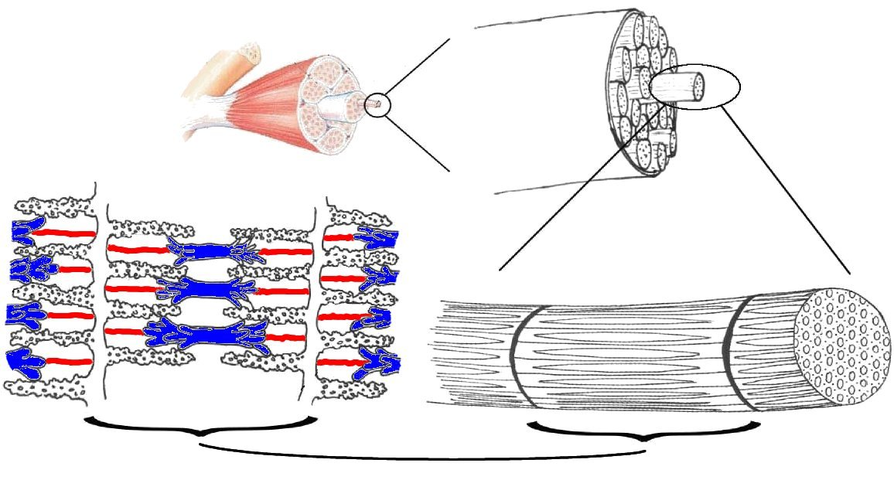
Adapted from Wikipedia
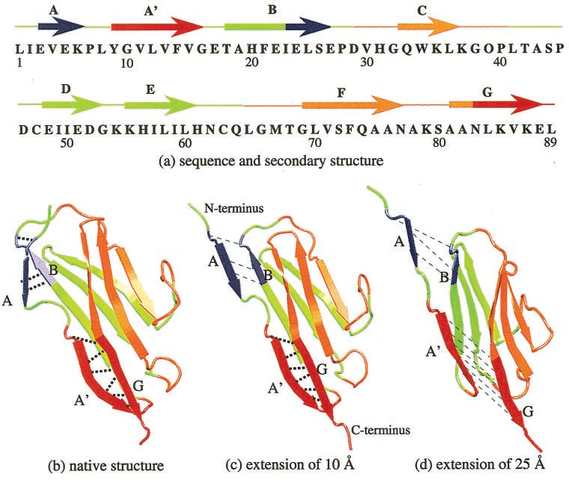
Proteins: Titin's I27
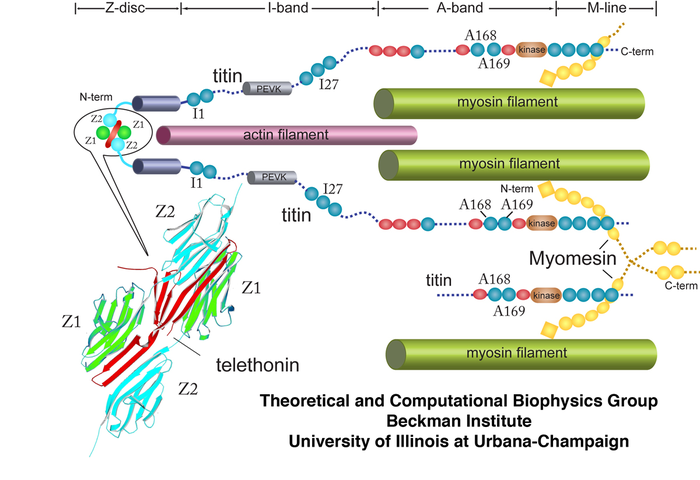
Proteins: I27
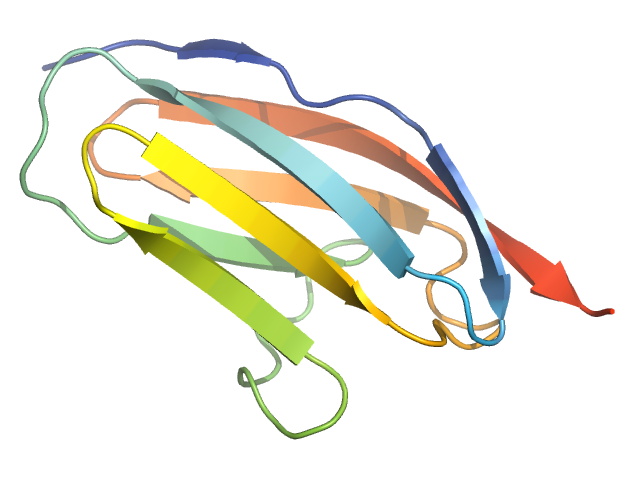
Proteins: What's the problem?
MHHHHHHSSLIEV EKPLYGVEVFVGE TAHFEIELSEPDV HGQWKLKGQPLTA SPDCEIIEDGKKH ILILHNCQLGMTG EVSFQAANAKSAA NLKVKEL |
→ |

|
|
|
||
Open source SMFS
Atomic force microscopy
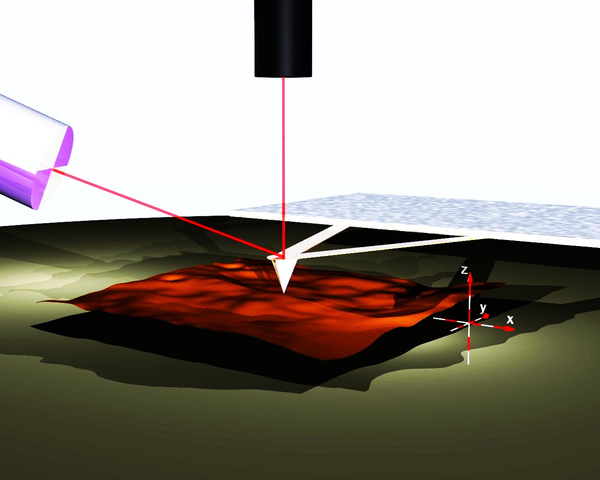
AFM: Cantilever geometry


Olympus TR800PSA
, images from
Asylum Research
We use the thinner
TR400PSA
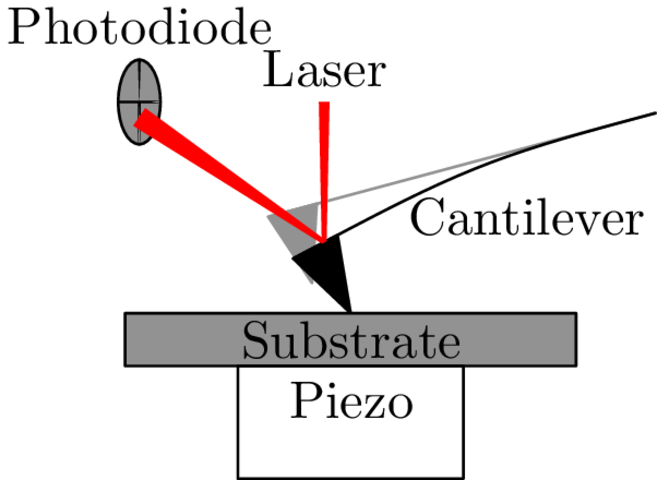
AFM: Laser deflection
AFM: Piezo positioning
The piezoelectric effect
Lead zirconium titanate (PZT) from Wikipedia
AFM: Tubular piezos
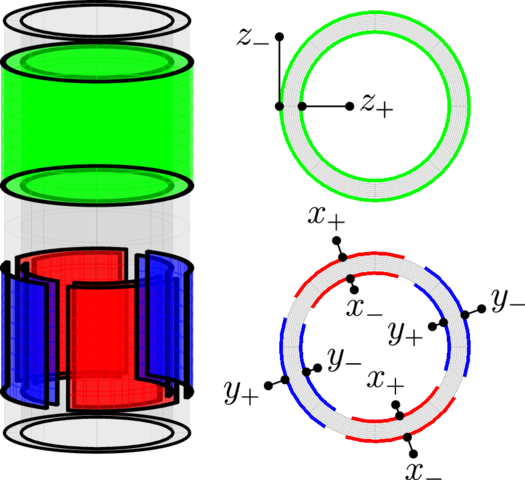
Open source SMFS
Single molecule force spectroscopy
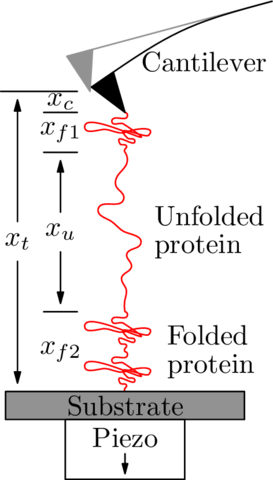
SMFS: Sawtooth curve
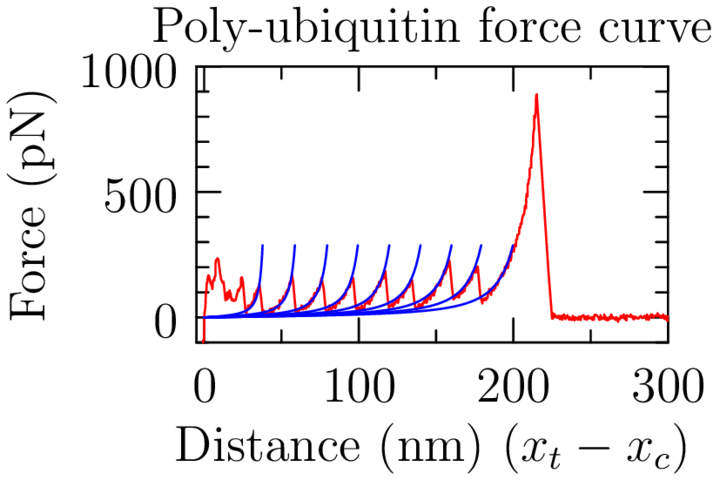
SMFS: What's going on?
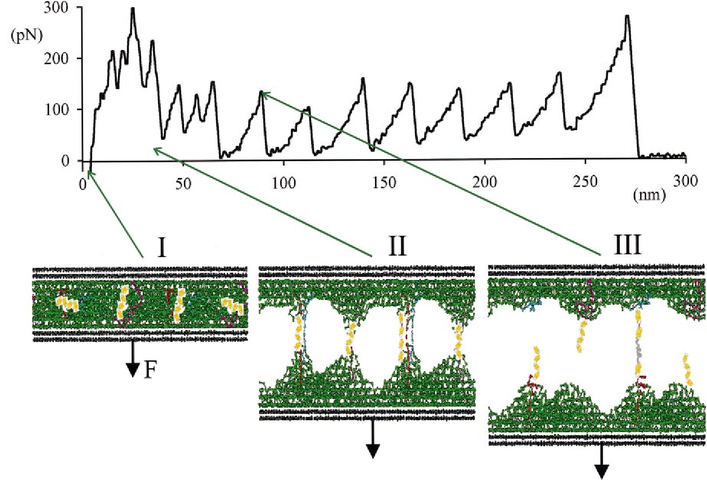
Carrion-Vazquez, et al., 2000; adapted from Baljon and Robbins, 1996
SMFS: Unfolding one domain
Open source SMFS
Experiment control
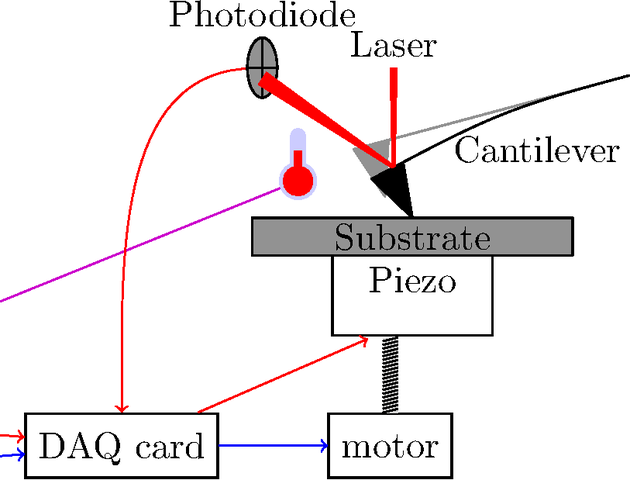
Control: Quick-and-dirty
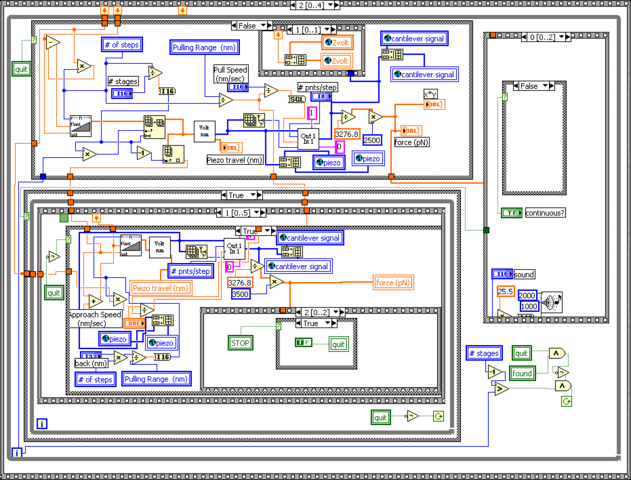
Control: My modular stack
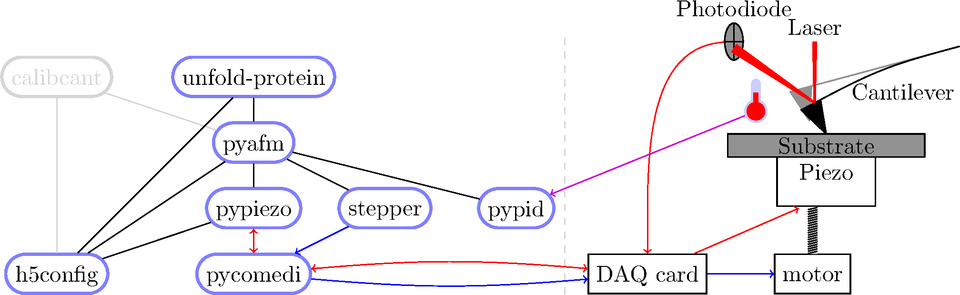
Open source: Existing layers
 |
||||
| Linux | GNU | Gentoo | Python | SciPy |
| Comedi | matplotlib | pymodbus | Cython | NumPy |
| h5py | … |
Open source: Teamwork

Control: Example code
class Unfolder (object):
…
def run(self):
"""Approach-bind-unfold-save[-plot] cycle.
"""
ret = {}
ret['timestamp'] = _email_utils.formatdate(localtime=True)
ret['temperature'] = self.afm.get_temperature()
ret['approach'] = self._approach()
self._bind()
ret['unfold'] = self._unfold()
self._save(**ret)
if _package_config['matplotlib']:
self._plot(**ret)
return ret
Archival: HDF5 and h5config
GROUP "/" GROUP "approach" … GROUP "config" GROUP "afm" … GROUP "approach" … DATASET "bind time" … GROUP "unfold" … DATASET "velocity" GROUP "environment" DATASET "temperature" DATASET "timestamp" … GROUP "unfold" DATASET "deflection" DATASET "frequency" DATASET "z"
Archival: Version control
commit 32bfbf98d79c73eba50b77d0917df100e0e33bcf Author: W. Trevor King <wking@tremily.us> Date: Fri Jan 18 22:54:49 2013 -0500 afm: Optionally return stepper_approach data with `record_data` Sometimes these approach curves are pretty funky, so I'll start recording them by default in calibcant-calibrate.py. diff --git a/pyafm/afm.py b/pyafm/afm.py index 60741c6..e76b118 100644 --- a/pyafm/afm.py +++ b/pyafm/afm.py @@ -460,10 +460,11 @@ class AFM (object): _LOG.warn(e) raise e - def stepper_approach(self, target_deflection): + def stepper_approach(self, target_deflection, record_data=None): …
Open source SMFS
Cantilever calibration

Calibration: Geometry


Olympus TR800PSA
, images from
Asylum Research
We use the thinner
TR400PSA
Calibration: Equipartition
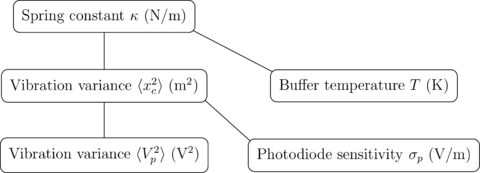
|
The average spring energy is where is Boltzmann's constant and is the temperature. |
Calibration: Vibration
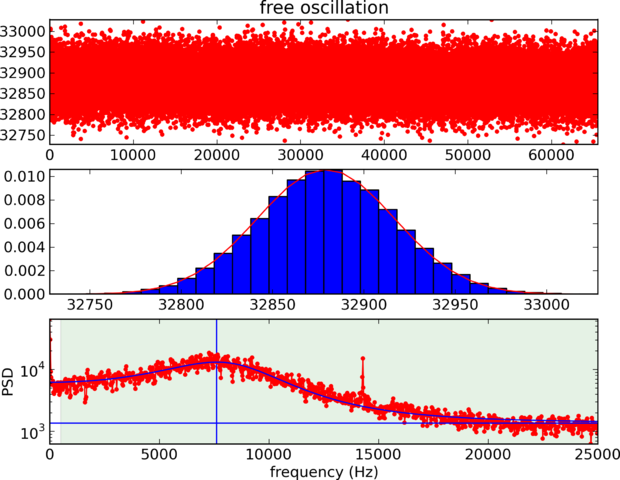
Calibration: Photodiode calibration
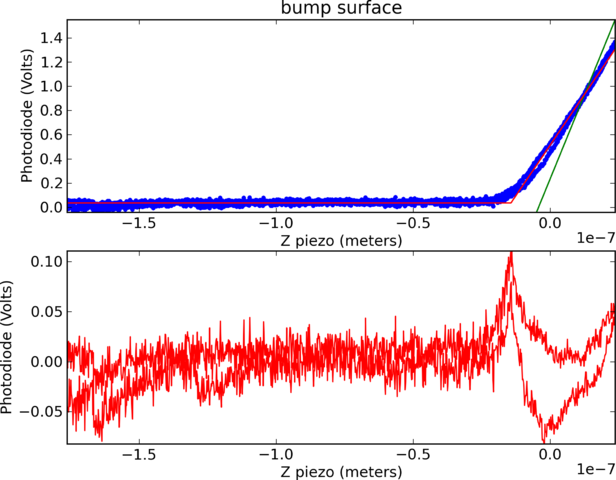
Calibration: Results
Calibration: Stability
| Quant. | Units | Day 1 | Day 2 | ||||
|---|---|---|---|---|---|---|---|
| K | 296.30 | ±0.02 | 294.27 | ±0.02 | |||
| mV/nm | 46.2 | ±0.8 | 41.3 | ±0.2 | |||
| mV | 108 | ±1 | 105 | ±2 | |||
| pN/nm | 67 | ±2 | 66 | ±2 | |||
Calibration: Inconsistency
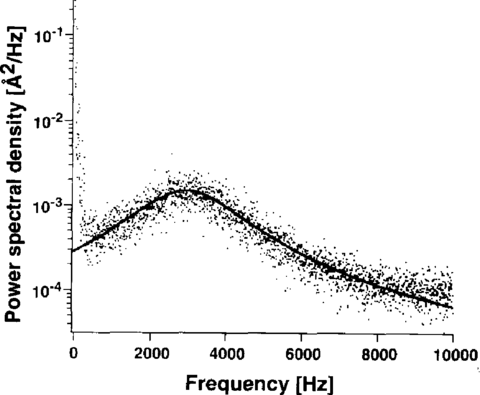
|
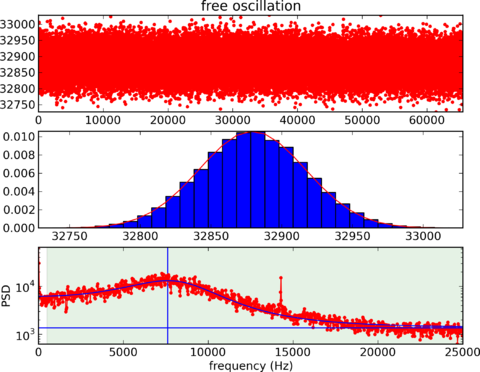
|
Open source SMFS
Monte Carlo unfolding simulations
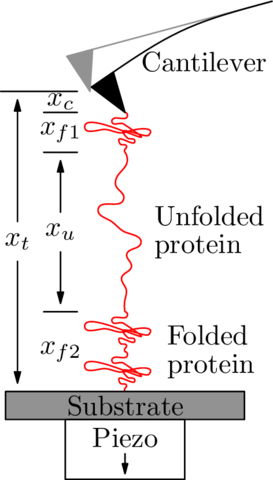
|
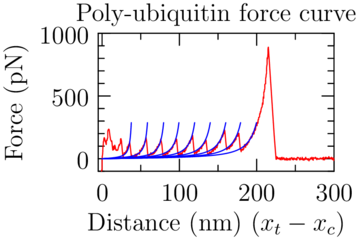
|
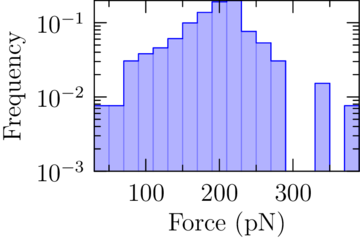 |
Hooke: Experimental histograms


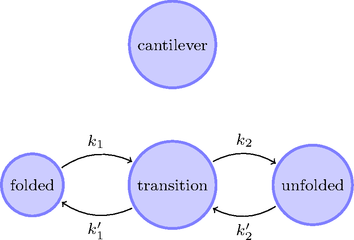
Sawsim: State model

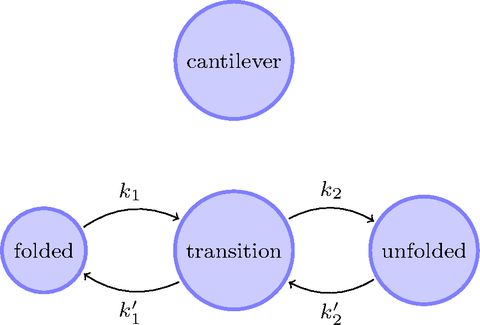
My simulation framework.
Sawsim: Simulation loop
- Calculate piezo-induced gap
- Find tension model parameters for each state
- Distribute per-state stretching (, , …) to balance the tension
- Calculate the transition rates between states
- Roll the dice to determine if transitions take place as you step forward in time
Sawsim: Monte Carlo
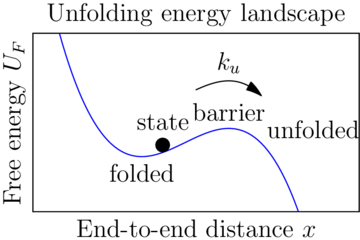
Sawsim: Unfolding models

|
|
|
|
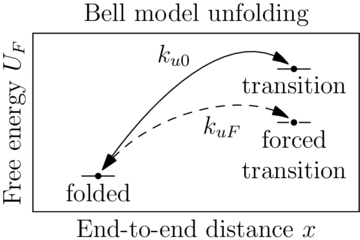
Sawsim: Kramers' model

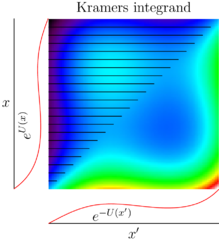
Sawsim: Tension models

|
|
|

|
Sawsim: Fitting models
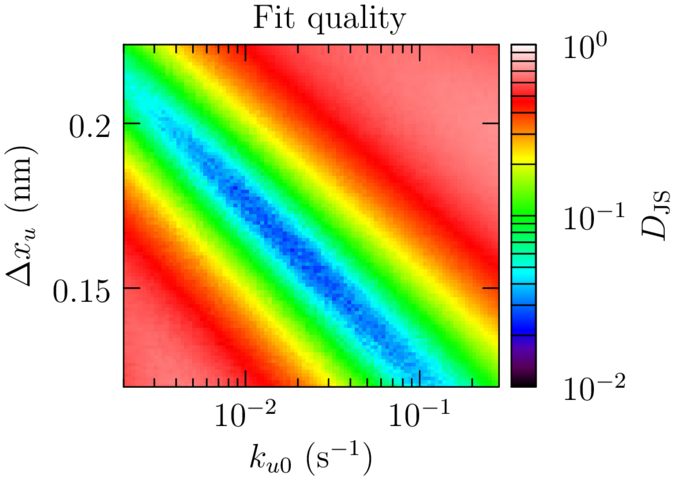
Open source SMFS
Unfolding in salty buffers

Salt: Glutamic acid
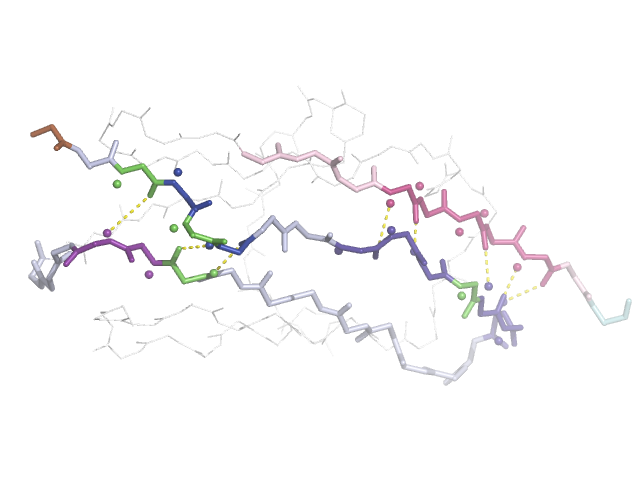
Salt: Reduced stability in CaCl₂
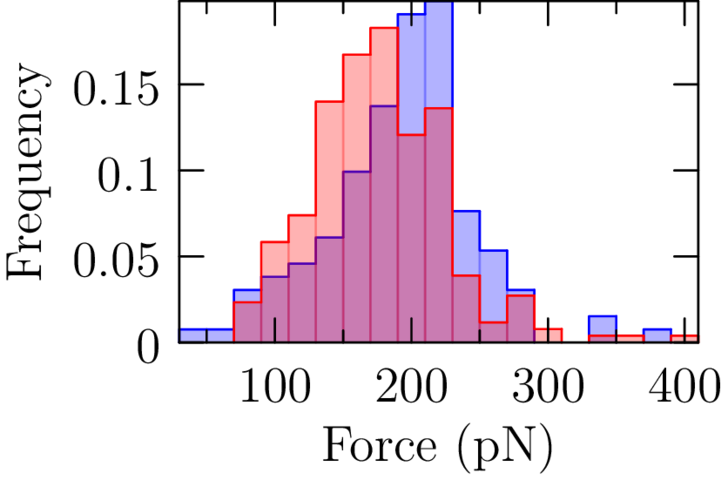
Salt: Sawsim fits
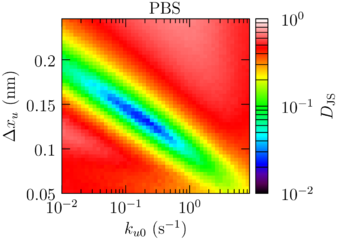
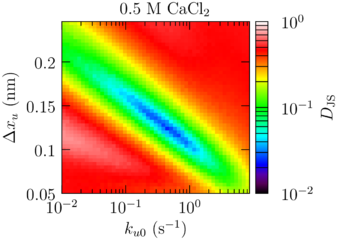
| Buffer | (Å) | (s) |
|---|---|---|
| PBS | 1.32 | 0.222 |
| PBS + 0.5 M CaCl₂ | 1.23 | 0.450 |
Ca²⁺ radius ∼1.1 Å, H-bond ∼2 Å.
Open source SMFS
Conclusions: Salt
- We see the expected destabilizing effect of CaCl₂
- Future work:
- More contextual data!
- Pulling speeds
- Salt concentrations (physiological levels)
- Salt species
- Mutated proteins?
- Is glutamic acid special?
- More contextual data!
Conclusions: Hardware
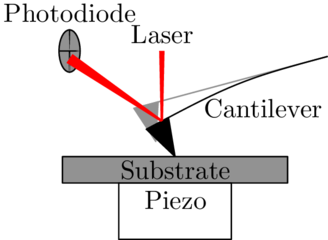
|
Open source AFM automation is possible, even with older hardware originally designed for more manual operation. |
Future: Hardware

|
For automatic control, it would be nice to have…
|
Conclusions: Software
New open source software:
Future: Software
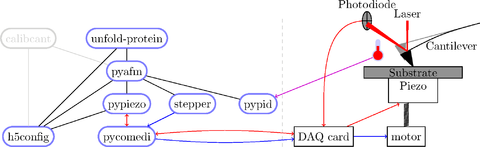
Everything works for me, and I expect it will work for others… but no software (except maybe TeX) is without bugs. Testers welcome!
The End
Thanks!
|
|
|